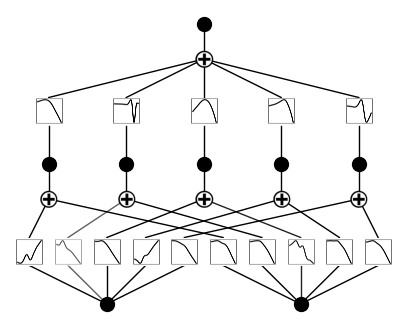
API 3: Extracting activation functions
The KAN diagrams give intuitive illustration, but sometimes we may also want to extract the values of activation functions for more quantitative tasks. Using the indexing convention introduced in the indexing notebook, each edge is indexed as \((l,i,j)\), where \(l\) is the layer index, \(i\) is the input neuron index, and \(j\) is output neuron index.
from kan import *
import matplotlib.pyplot as plt
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print(device)
# create a KAN: 2D inputs, 1D output, and 5 hidden neurons. cubic spline (k=3), 5 grid intervals (grid=5).
model = KAN(width=[2,5,1], grid=3, k=3, seed=1, device=device)
x = torch.normal(0,1,size=(100,2)).to(device)
model(x)
model.plot(beta=100)
cuda
checkpoint directory created: ./model
saving model version 0.0
l = 0
i = 0
j = 3
x, y = model.get_fun(l,i,j)
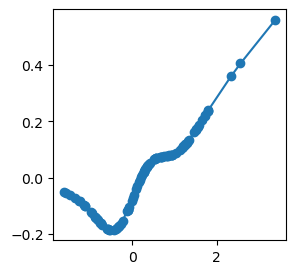
If we are interested in the range of some activation function, we can use get_range.
model.get_range(l,i,j)
x range: [-1.61 , 3.38 ]
y range: [-0.19 , 0.56 ]
(array(-1.6111118, dtype=float32),
array(3.38374, dtype=float32),
array(-0.18606013, dtype=float32),
array(0.5614974, dtype=float32))