API 2: Plotting
Initialize KAN and create dataset
from kan import *
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
print(device)
# create a KAN: 2D inputs, 1D output, and 5 hidden neurons. cubic spline (k=3), 5 grid intervals (grid=5).
model = KAN(width=[2,5,1], grid=3, k=3, seed=1, device=device)
# create dataset f(x,y) = exp(sin(pi*x)+y^2)
f = lambda x: torch.exp(torch.sin(torch.pi*x[:,[0]]) + x[:,[1]]**2)
dataset = create_dataset(f, n_var=2, device=device)
dataset['train_input'].shape, dataset['train_label'].shape
cuda
checkpoint directory created: ./model
saving model version 0.0
(torch.Size([1000, 2]), torch.Size([1000, 1]))
Plot KAN at initialization
# plot KAN at initialization
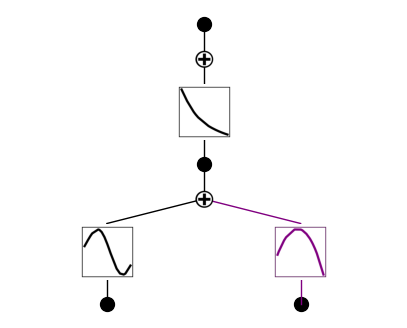
model(dataset['train_input']);
model.plot(beta=100)
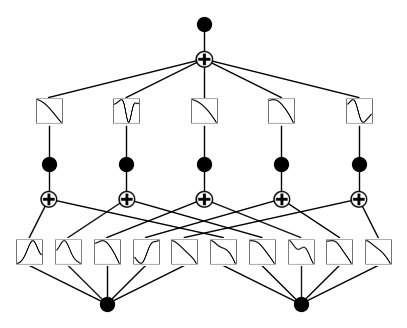
# if you want to add variable names and title
model.plot(beta=100, in_vars=[r'$\alpha$', 'x'], out_vars=['y'], title = 'My KAN')
Train KAN with sparsity regularization
# train the model
model.fit(dataset, opt="LBFGS", steps=20, lamb=0.01);
| train_loss: 5.20e-02 | test_loss: 5.35e-02 | reg: 4.93e+00 | : 100%|█| 20/20 [00:03<00:00, 5.22it
saving model version 0.1
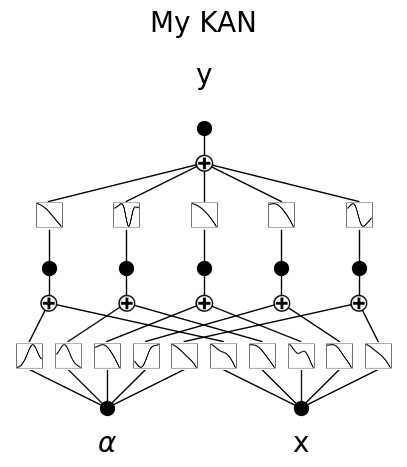
\(\beta\) controls the transparency of activations. Larger \(\beta\) => more activation functions show up. We usually want to set a proper beta such that only important connections are visually significant. transparency is set to be \({\rm tanh}(\beta \phi)\) where \(\phi\) is the scale of the activation function (metric=‘forward_u’), normalized scale (metric=‘forward_n’) or the feature attribution score (metric=‘backward’). By default \(\beta=3\) and metric=‘backward’.
model.plot()
model.plot(beta=100000)
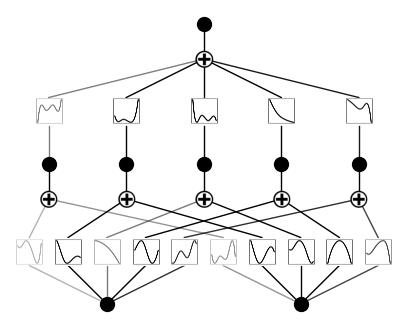
model.plot(beta=0.1)
plotting with different metrics: ‘forward_n’, ‘forward_u’, ‘backward’
model.plot(metric='forward_n', beta=100)
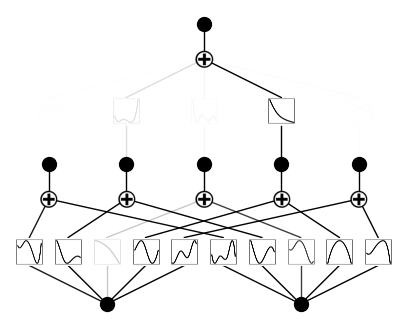
model.plot(metric='forward_u', beta=100)
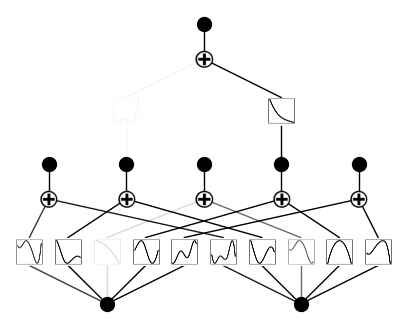
model.plot(metric='backward', beta=100)
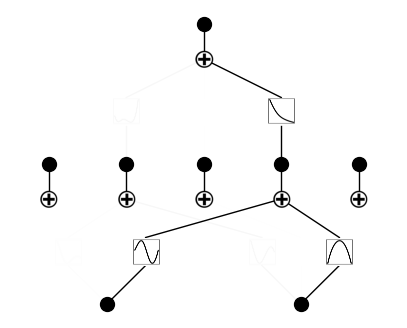
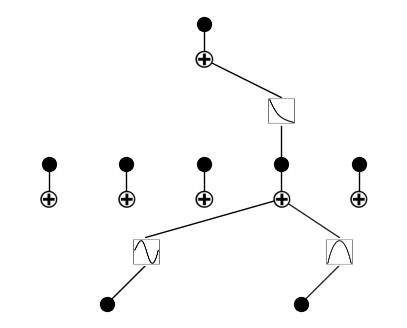
Remove insignificant neurons
model = model.prune()
model.plot()
saving model version 0.2
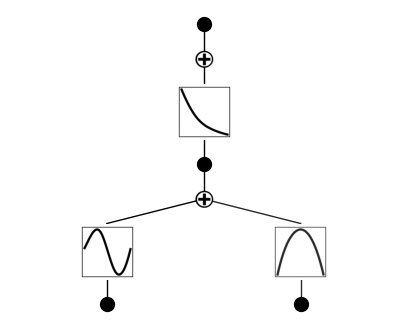
Resize the figure using the “scale” parameter. By default: 0.5
model.plot(scale=0.5)
model.plot(scale=0.2)
model.plot(scale=2.0)
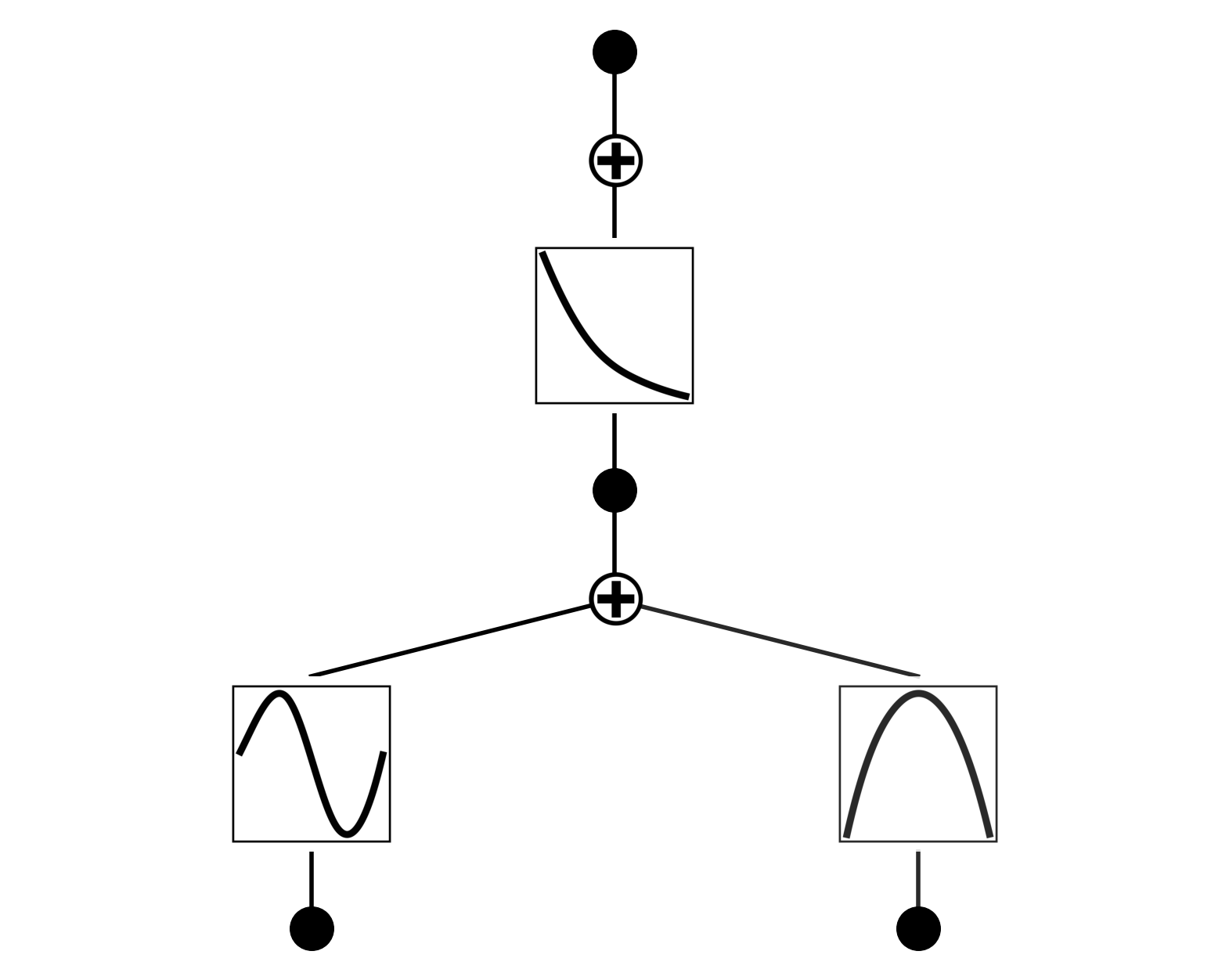
If you want to see sample distribution in addition to the line, set “sample=True”
model.plot(sample=True)
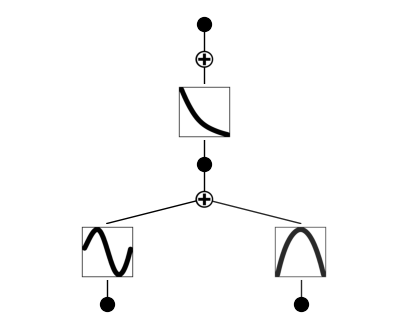
The samples are more visible if we use a smaller number of samples
model.get_act(dataset['train_input'][:20])
model.plot(sample=True)
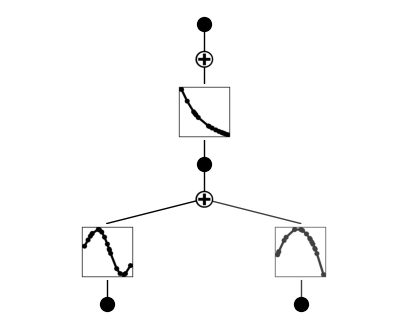
If a function is set to be symbolic, it becomes red
model.fix_symbolic(0,1,0,'x^2')
r2 is 0.9992202520370483
saving model version 0.3
tensor(0.9992, device='cuda:0')
model.plot()
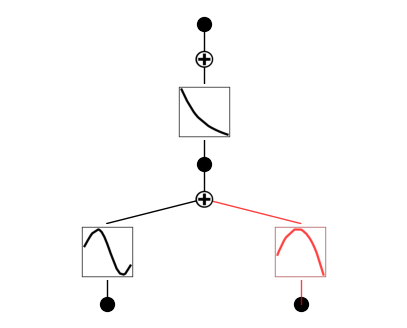
If a function is set to be both symbolic and numeric (its output is the addition of symbolic and spline), then it shows up in purple
model.set_mode(0,1,0,mode='ns')
model.plot(beta=100)